Recently I had the pleasure of teaching at CEU (see in-progress course materials here). One of the hardest concepts to explain turned out to be the role of validation and test sets.
In short, a usual process for building and evaluating predictive models is the following. We divide our data into three parts. On the first part (“training set”) we are estimating the candidate models, on the second (“validation set”) we are comparing their performances and select the best one. Finally, on the third part (“test set”) we are evaluating the performance of the best model.
Why do we need the final evaluation on the test set? After all, we are already calculating the performance measure of models on the validation set. It is an independent data set from the training sample, so we are guarded against overfitting, right?
On swimming and predictive models
A student of mine came up with a very clever analogy between predictive modelling and swimming competitions. Swimmers train a lot in their home countries, day after day and their times are recorded. However, in order to be able to fairly compare their performances and to choose the best swimmer, we hold the Olympic Games where the winner is claimed to be the best in the world. Once predictive models have been trained, we use validation set as the Olympic Games where their performance can be compared.
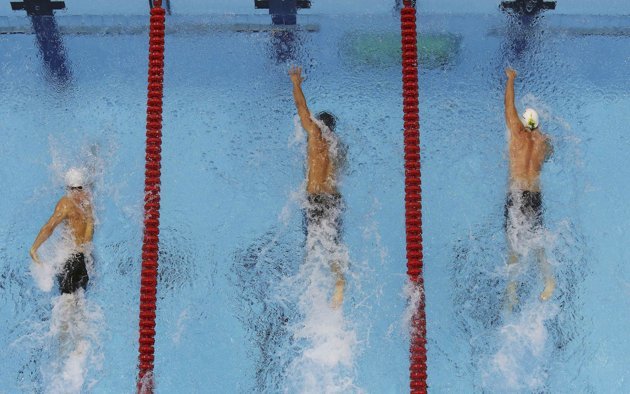
In sports, that’s the end of the story. However, with predictive models we want to have a performance measure that shows how the model is going to perform in general, for all subsequent new cases. To continue the analogy, in machine learning we select the Olympic gold medalist, however, we want to have a clear picture about how she would perform in a lifelong competition made up of many different races.
Often times small peculiarities of competitions can influence who comes out as the best even in the Olympic finals. This means that overfitting can happen even in the finals: there is a chance that the winner only had a lucky day and that is why she comes out as the best on that day, in that swimming pool, etc. Selecting her as the winner and claiming the winner time to be her overall performance would be confounded by these lucky circumstances that positively influenced her achievement.
We want to make sure that when we select one model that we are going to use we are not fooled by such peculiarities that are only characterizing the Olympic finals. By calculating performance of the selected model on the test set we are getting an unbiased measure of how the model would perform in the lifelong competition.
Illustration
We can illustrate the issue by performing a simple prediction task where, for simplicity, we compare models of equal performance. We try to predict diabetes among Pima people using 50 models. These are models of one variable that do not have any relation to the outcome, hence their explanatory power is zero. Hence, no matter which model we select, its performance measured by AUC should be close to 0.5.
library(purrr)
library(data.table)
library(mlbench)
library(ROCR)
set.seed(1234)
data("PimaIndiansDiabetes")
data <- data.table(PimaIndiansDiabetes)
data <- data[, .(diabetes)]
num_models <- 50
variable_names <- map_chr(1:num_models, ~{ paste0("X_", .x) })
# create the variables that will be used to explain the outcome
# they are completely random and independent from the outcome
walk(variable_names,
~{
varname <- .x
data[, (varname) := rnorm(nrow(data))]
}
)Let’s create train, validation and test sets.
ix_train <- sample(1:nrow(data), as.integer(nrow(data) * 0.5))
data_train <- data[ix_train,]
data_validation_test <- data[-ix_train,]
ix_validation <- sample(1:nrow(data_validation_test),
as.integer(nrow(data_validation_test) * 0.5))
data_validation <- data_validation_test[ix_validation,]
data_test <- data_validation_test[-ix_validation,]A simple logistic model is used for prediction: for each of the 50 variables we estimate a one-variable regression.
models <- map(variable_names,
~ {
model <- glm(formula(paste0("diabetes ~ ", .x)),
data = data_train,
family = binomial)
}
)Let us calculate the AUC of these models on the validation set and see how the best model performs.
auc_scores <- map_dbl(
models,
~ {
predicted_score <- predict(.x,
newdata = data_validation,
type = "response")
performance(prediction(predicted_score,
data_validation[["diabetes"]]),
"auc")@y.values[[1]]
}
)
max(auc_scores)## [1] 0.6095323The maximum AUC is well-above the expected 0.5. The best performing model on the validation set seemingly has a performance that is way better than random. However, this is only purely by chance and is a sign of overfitting for the validation set.
We can see it by using the test set as a final evaluation of the selected “best” model where the performance is much closer to the expected 0.5.
best_model <- models[[which.max(auc_scores)]]
predicted_score <- predict(best_model,
newdata = data_test,
type = "response")
performance(prediction(predicted_score, data_test[["diabetes"]]),
"auc")@y.values[[1]] ## [1] 0.4916996Conclusion
This extreme example shows why it is essential to have a set of data used only to evaluate the final model, without having been used in any ways for model building or selection.
The problem that we solve with having separate validation and test sets is very much the same as multiple testing. If we perform many hypothesis tests and look at their p-values in isolation, even if in all cases the null hypothesis is true, in some cases we are going to have very low p-values purely by chance.